- Главная
- Разное
- Дизайн
- Бизнес и предпринимательство
- Аналитика
- Образование
- Развлечения
- Красота и здоровье
- Финансы
- Государство
- Путешествия
- Спорт
- Недвижимость
- Армия
- Графика
- Культурология
- Еда и кулинария
- Лингвистика
- Английский язык
- Астрономия
- Алгебра
- Биология
- География
- Детские презентации
- Информатика
- История
- Литература
- Маркетинг
- Математика
- Медицина
- Менеджмент
- Музыка
- МХК
- Немецкий язык
- ОБЖ
- Обществознание
- Окружающий мир
- Педагогика
- Русский язык
- Технология
- Физика
- Философия
- Химия
- Шаблоны, картинки для презентаций
- Экология
- Экономика
- Юриспруденция
Crowdsourcing Biology: The Gene Wiki, BioGPS and GeneGames.org презентация
Содержание
- 1. Crowdsourcing Biology: The Gene Wiki, BioGPS and GeneGames.org
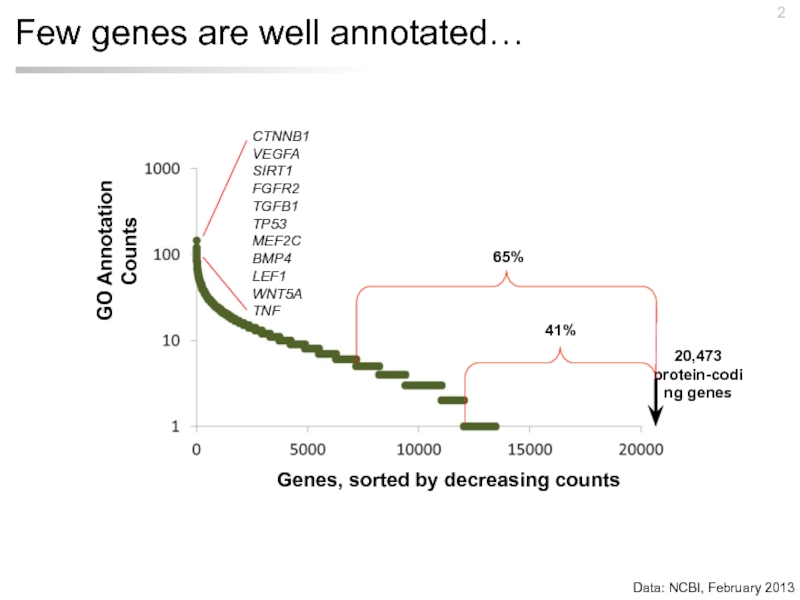
- 2. Few genes are well annotated… Data: NCBI, February 2013
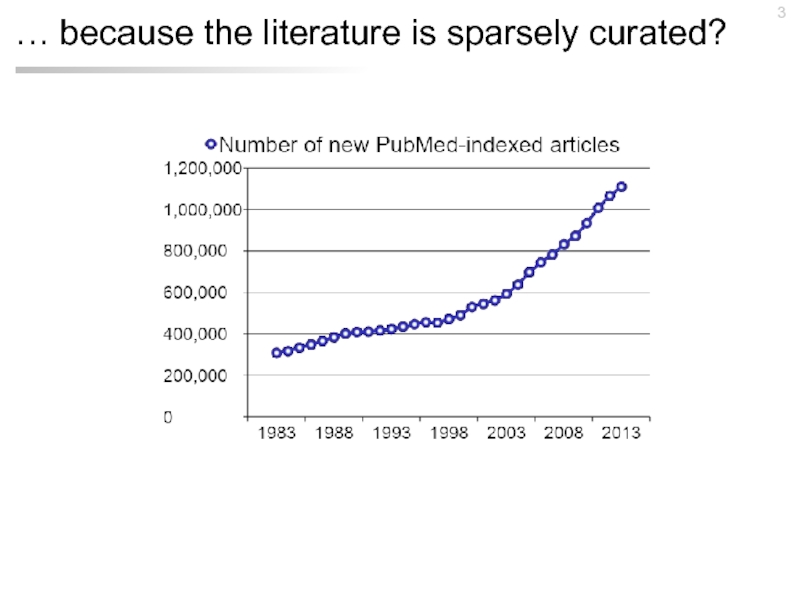
- 3. … because the literature is sparsely curated?
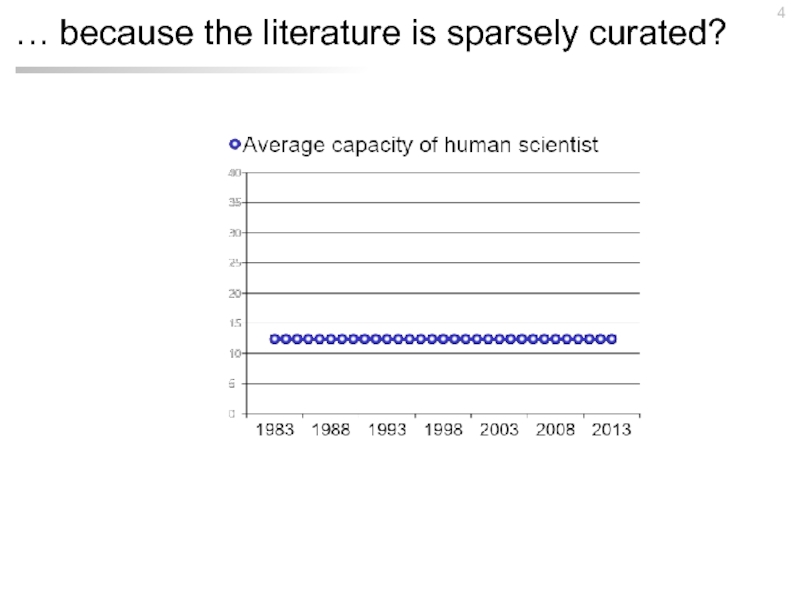
- 4. … because the literature is sparsely curated?
- 5. 311,696 articles (1.5% of PubMed) have been cited by GO annotations
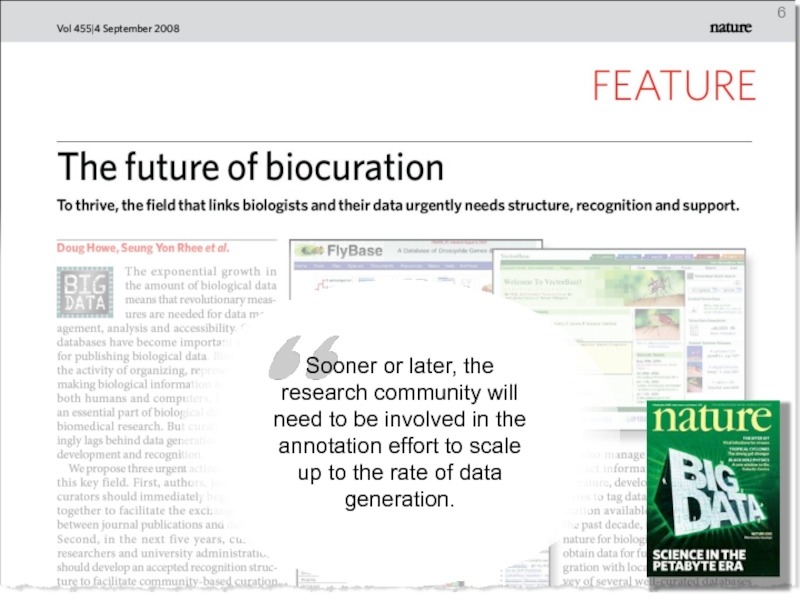
- 6. 0 Sooner or later, the research community
- 7. The Long Tail is a prolific source
- 8. Wikipedia is reasonably accurate
- 9. Wikipedia has breadth and depth http://en.wikipedia.org/wiki/Wikipedia:Size_comparisons, July 2008
- 10. We can harness the Long Tail of
- 11. From crowdsourcing to structured data
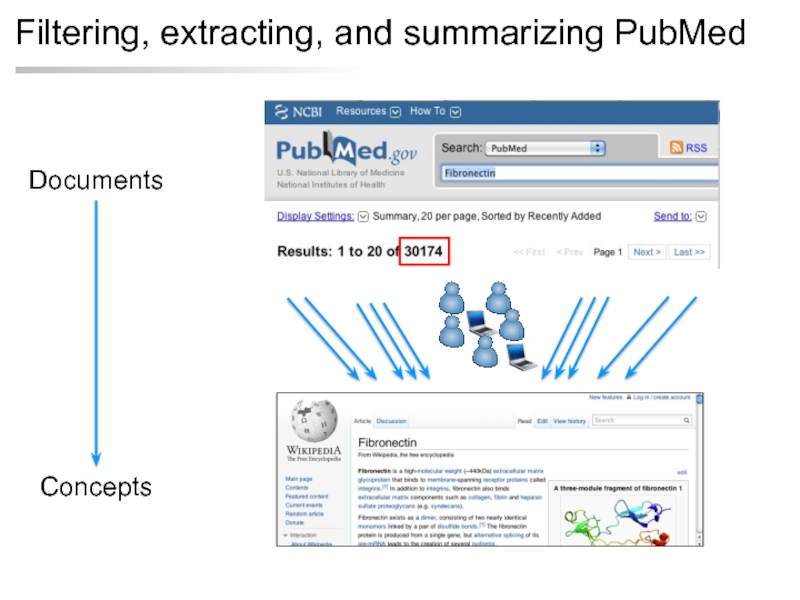
- 12. Filtering, extracting, and summarizing PubMed Documents Concepts Review article
- 13. Filtering, extracting, and summarizing PubMed Documents Concepts
- 14. Wiki success depends on a positive feedback
- 15. 10,000 gene “stubs” within Wikipedia Protein structure
- 16. Gene Wiki has a critical mass of
- 17. Gene Wiki has a critical mass of
- 18. A review article for every gene is
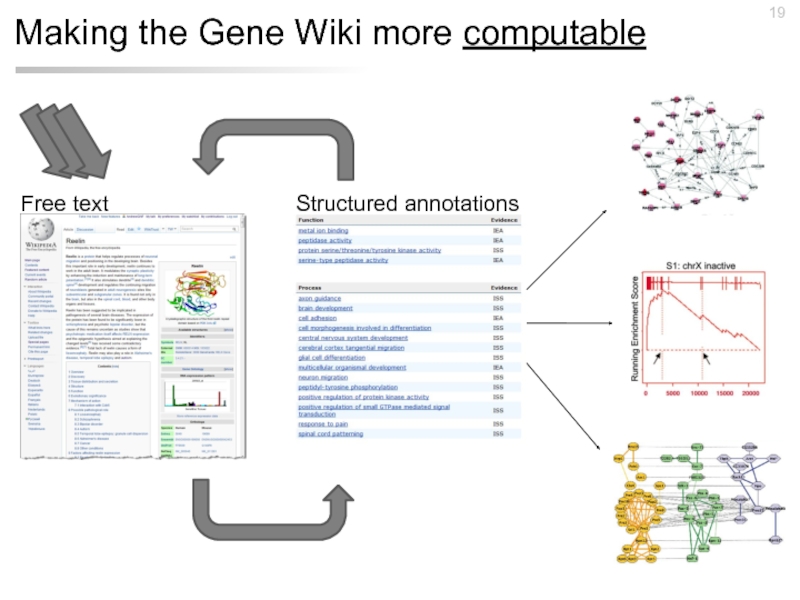
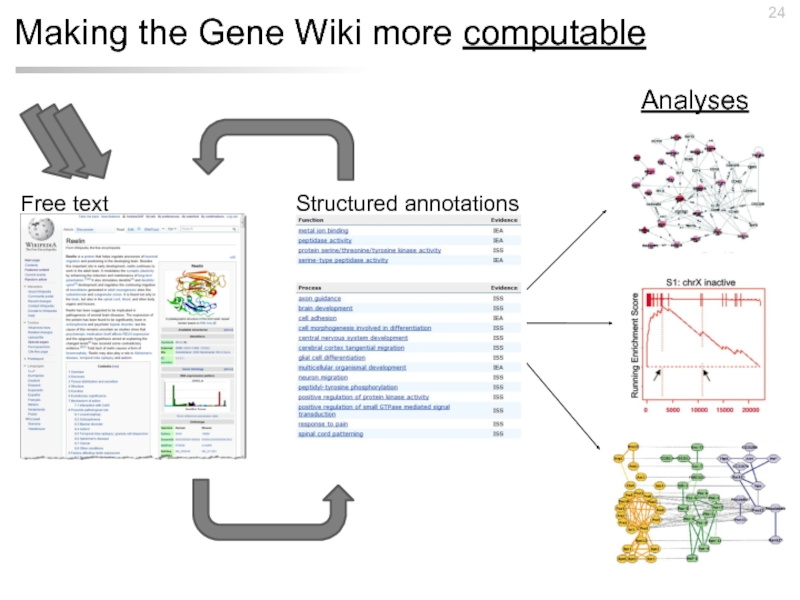
- 19. Making the Gene Wiki more computable Structured annotations Free text
- 20. Filling the gaps in gene annotation 6319 novel GO annotations 2147 novel DO annotations
- 21. Gene Wiki content improves enrichment analysis GO
- 22. Gene Wiki content improves enrichment analysis GO
- 23. Gene Wiki content improves enrichment analysis p-value
- 24. Making the Gene Wiki more computable Structured annotations Free text Analyses
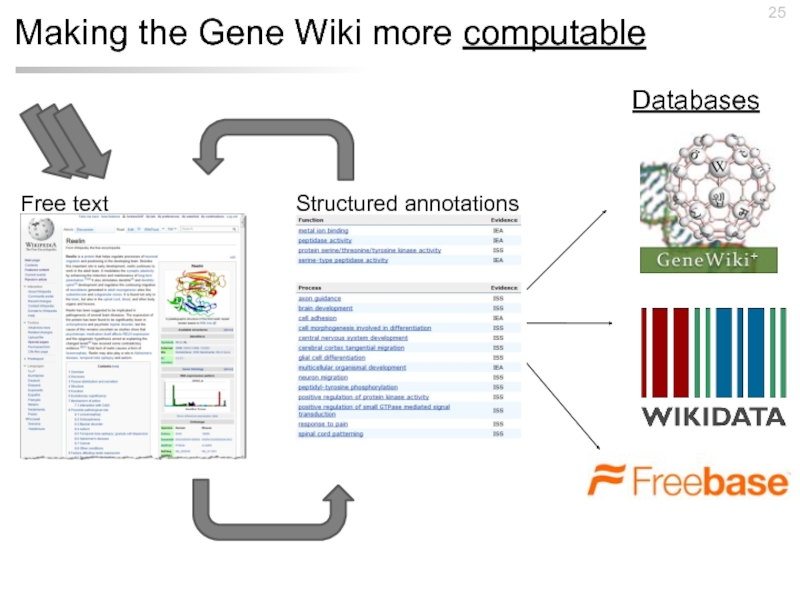
- 25. Making the Gene Wiki more computable Structured annotations Free text Databases
- 26. Expansion through outreach and incentives
- 27. Cardiovascular Gene Wiki Portal CAMK2D -- CaM
- 28. The Long Tail of scientists
- 29. From crowdsourcing to structured data
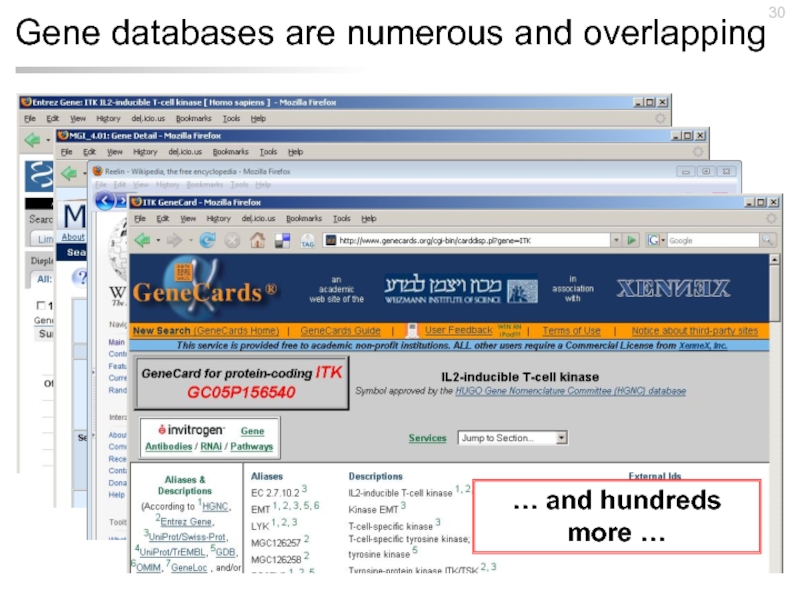
- 30. Gene databases are numerous and overlapping … and hundreds more …
- 31. Why is there so much redundancy? Users
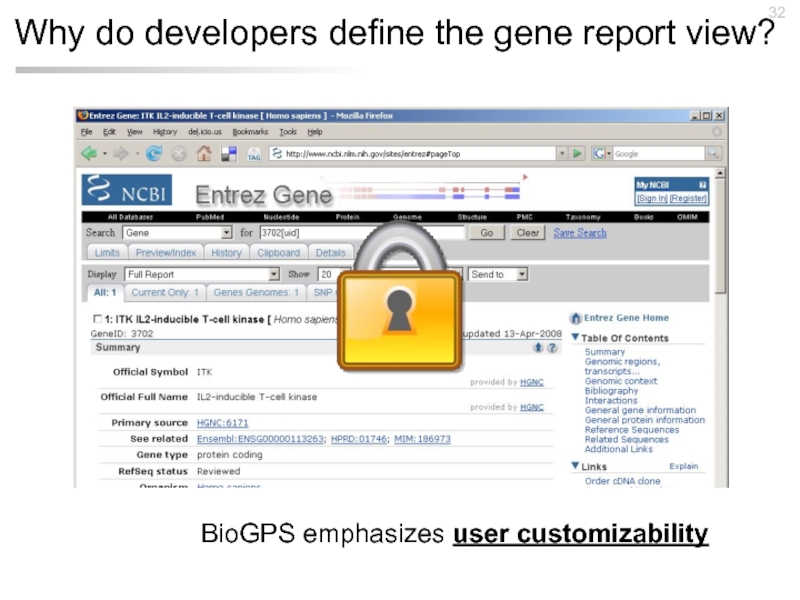
- 32. Why do developers define the gene report view? BioGPS emphasizes user customizability
- 33. Community extensibility and user customizability
- 34. Utility: A simple and universal plugin interface
- 35. Utility: A simple and universal plugin interface
- 36. Utility: A simple and universal plugin interface
- 37. Utility: A simple and universal plugin interface
- 38. Utility: A simple and universal plugin interface
- 39. Utility: A simple and universal plugin interface
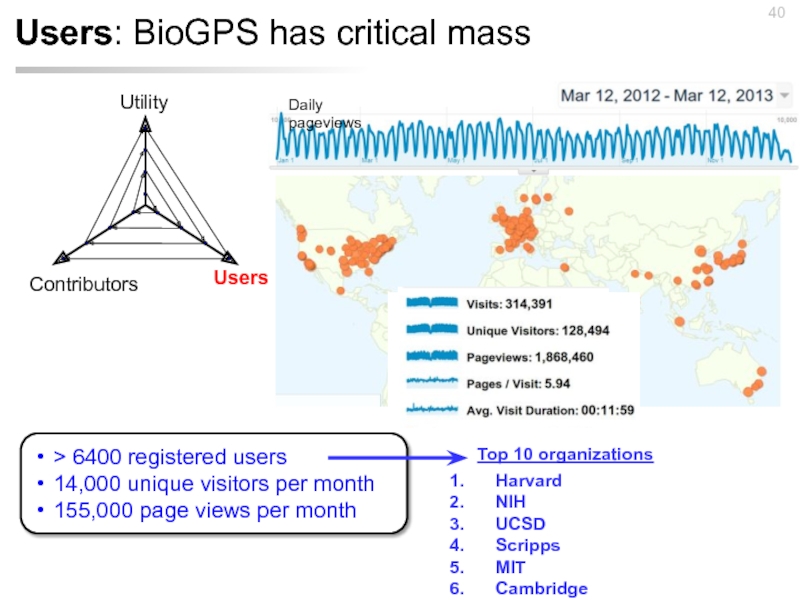
- 40. Users: BioGPS has critical mass Daily pageviews
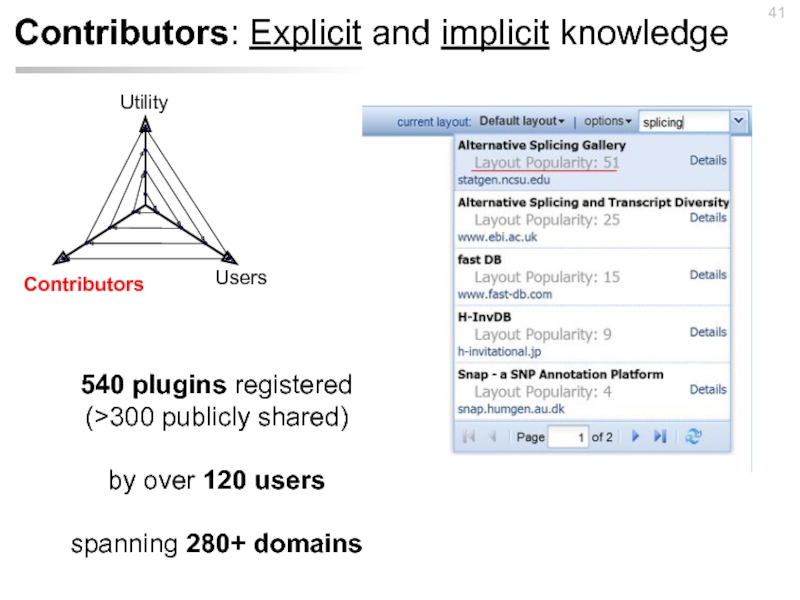
- 41. Contributors: Explicit and implicit knowledge 540 plugins
- 42. Gene Annotation Query as a Service http://mygene.info
- 43. The Long Tail of bioinformaticians can collaboratively build a gene portal.
- 44. From crowdsourcing to structured data
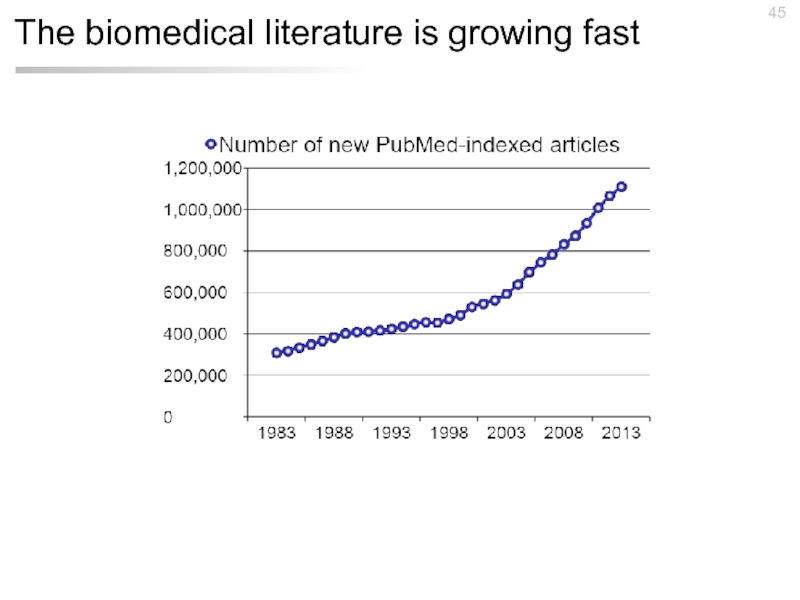
- 45. The biomedical literature is growing fast
- 46. Information Extraction Find mentions of high level
- 47. Disease mentions in PubMed abstracts NCBI Disease
- 48. Four types of disease mentions Specific Disease:
- 49. Question: Can a group of non-scientists collectively perform concept recognition in biomedical texts?
- 50. The Turk http://en.wikipedia.org/wiki/The_Turk
- 51. The Turk http://en.wikipedia.org/wiki/The_Turk
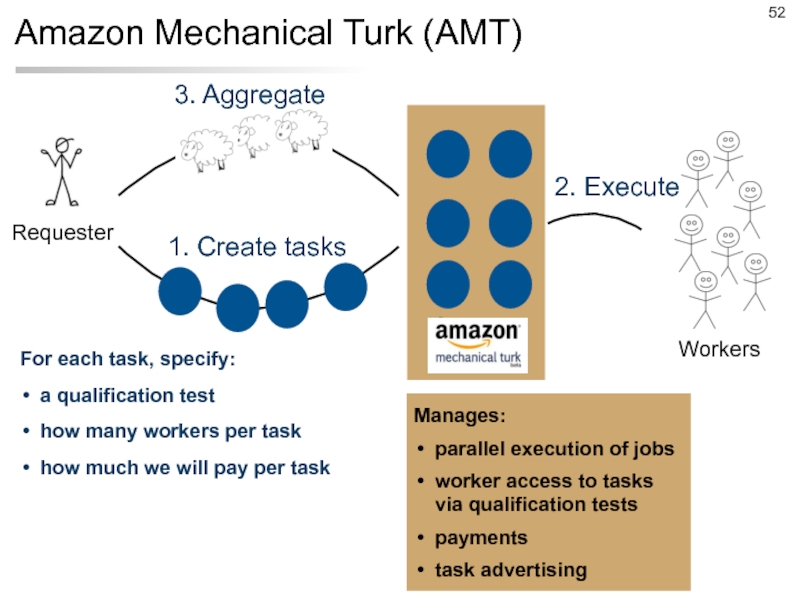
- 52. Amazon Mechanical Turk (AMT) For each task,
- 53. Instructions to workers Highlight all diseases and
- 54. Qualification test Test #1: “Myotonic dystrophy (
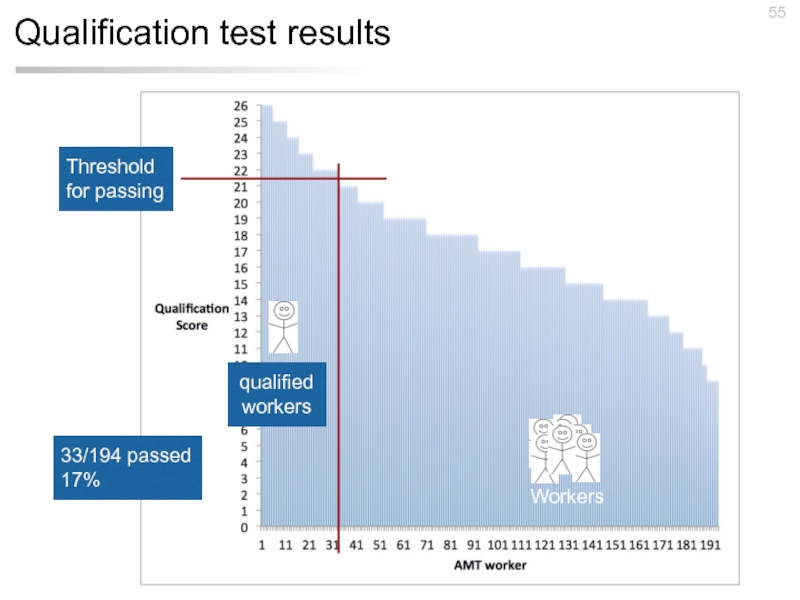
- 55. Qualification test results
- 56. Simple annotation interface Click to see instructions Highlight disease mentions
- 57. Experimental design Task: Identify the disease mentions
- 58. Aggregation function based on simple voting 1 or more votes (K=1) K=2 K=3 K=4
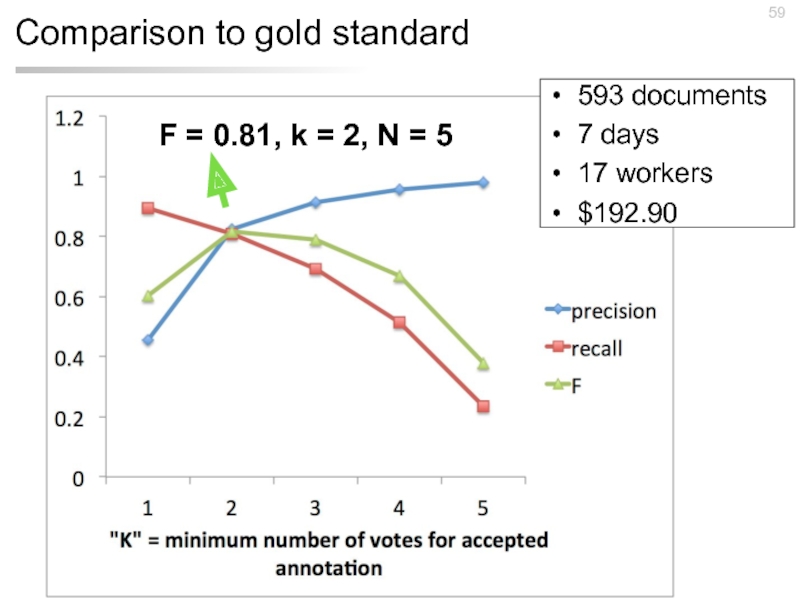
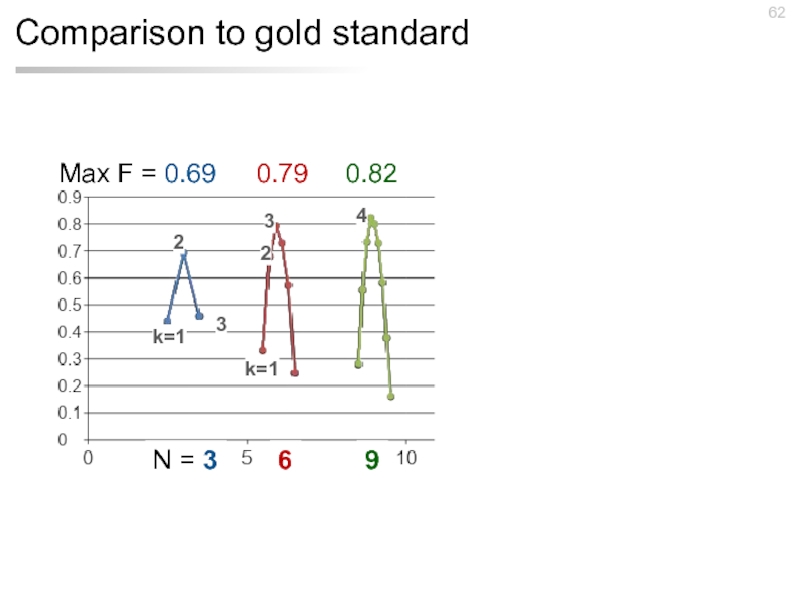
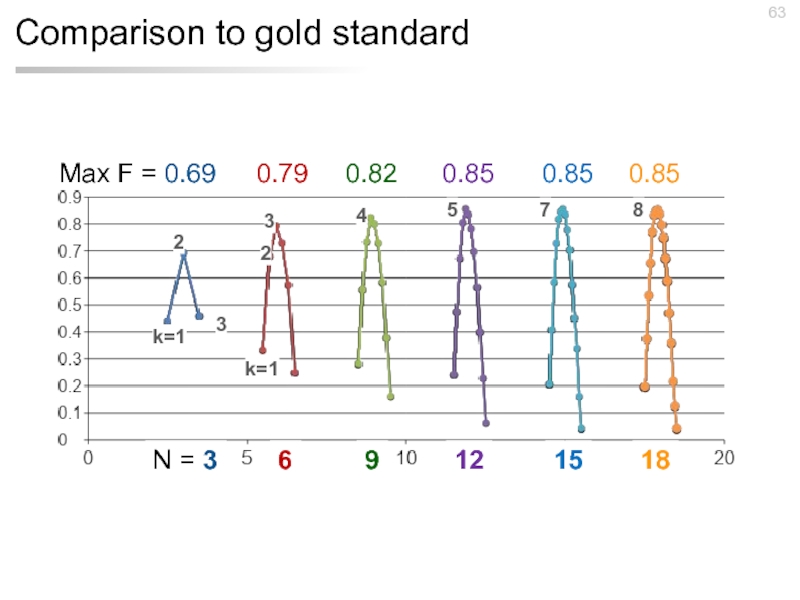
- 59. Comparison to gold standard 593 documents 7 days 17 workers $192.90
- 60. Comparison to gold standard Max F =
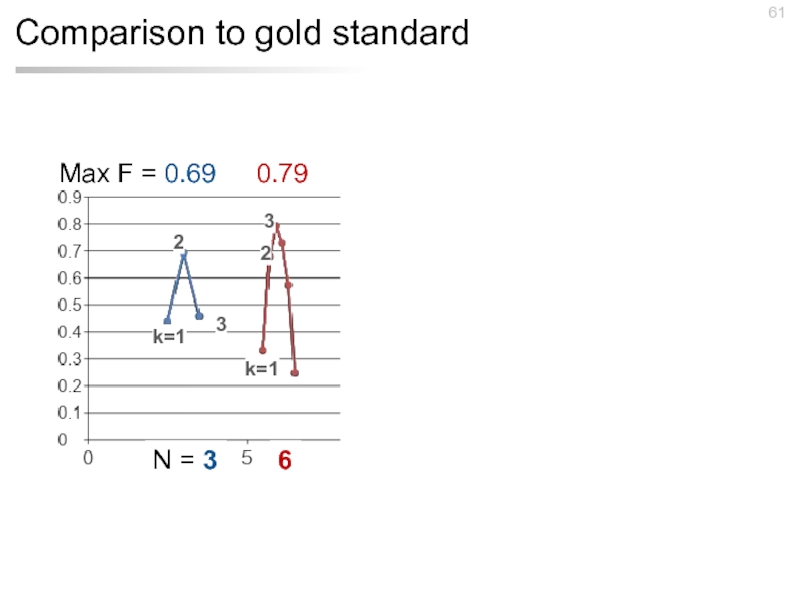
- 61. Comparison to gold standard Max F =
- 62. Comparison to gold standard Max F =
- 63. Comparison to gold standard Max F =
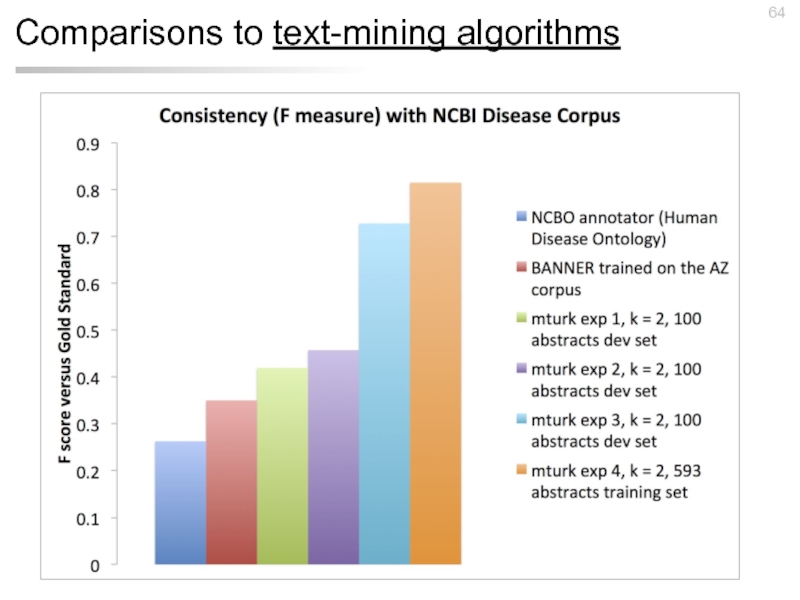
- 64. Comparisons to text-mining algorithms
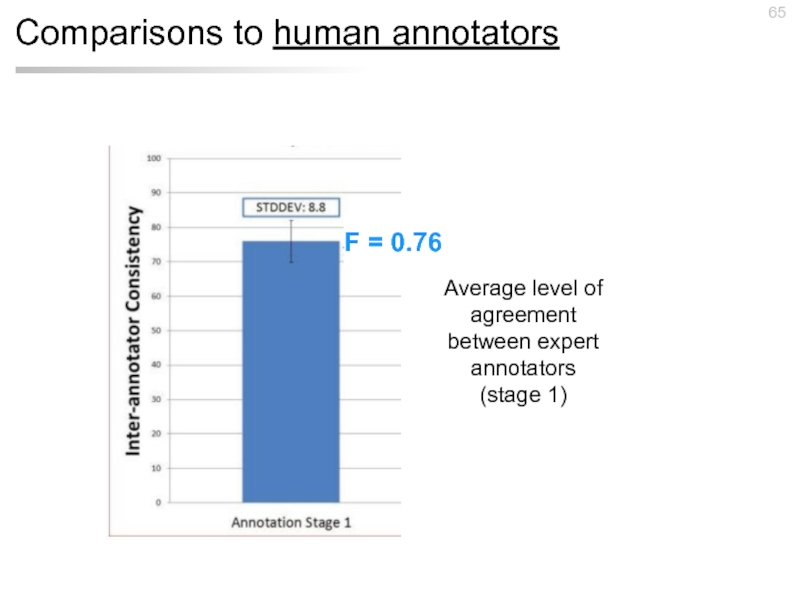
- 65. Comparisons to human annotators Average level of
- 66. Comparisons to human annotators F = 0.76
- 67. In aggregate, our worker ensemble is faster,
- 68. Information Extraction Find mentions of high level
- 69. Annotating the relationships This molecule inhibits the
- 70. Citizen Science at Mark2Cure.org
- 71. The Long Tail of citizen scientists can collaboratively annotate biomedical text.
- 72. Doug Howe, ZFIN John Hogenesch, U Penn
- 73. Related AMT work [1] Zhai et al
Слайд 1Crowdsourcing Biology: The Gene Wiki, BioGPS and GeneGames.org
May 14, 2014
CBIIT
Slides: slideshare.net/andrewsu
Citizen
Слайд 60
Sooner or later, the research community will need to be involved
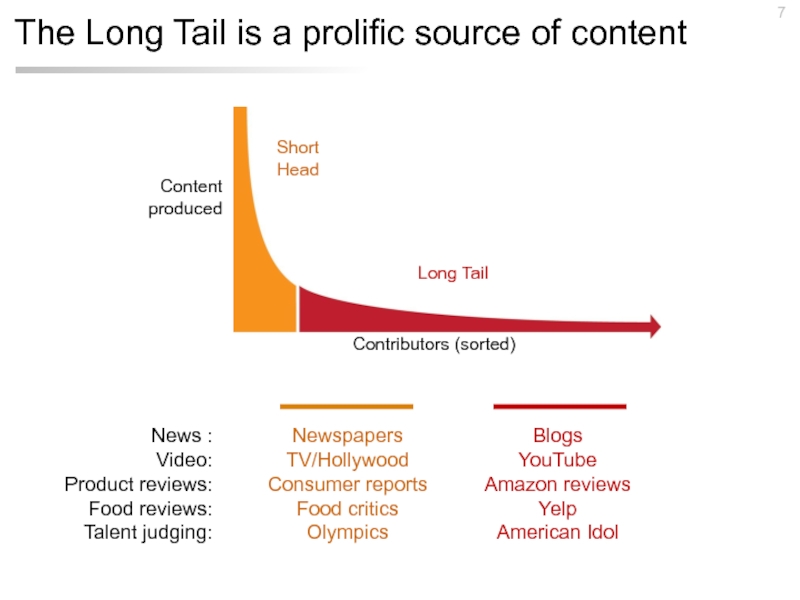
Слайд 7The Long Tail is a prolific source of content
News :
Video:
Product reviews:
Food
Talent judging:
Newspapers
TV/Hollywood
Consumer reports
Food critics
Olympics
Blogs
YouTube
Amazon reviews
Yelp
American Idol
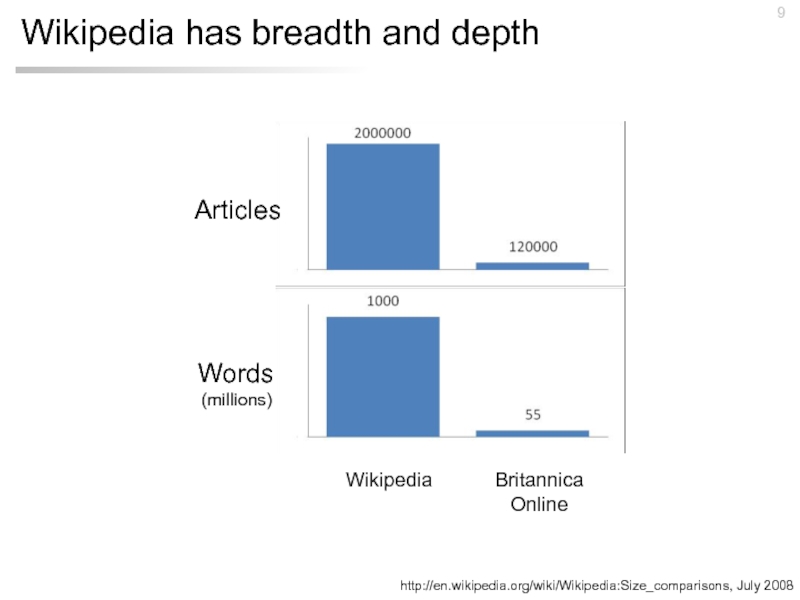
Слайд 9Wikipedia has breadth and depth
http://en.wikipedia.org/wiki/Wikipedia:Size_comparisons, July 2008
Слайд 10We can harness the Long Tail of scientists to directly participate
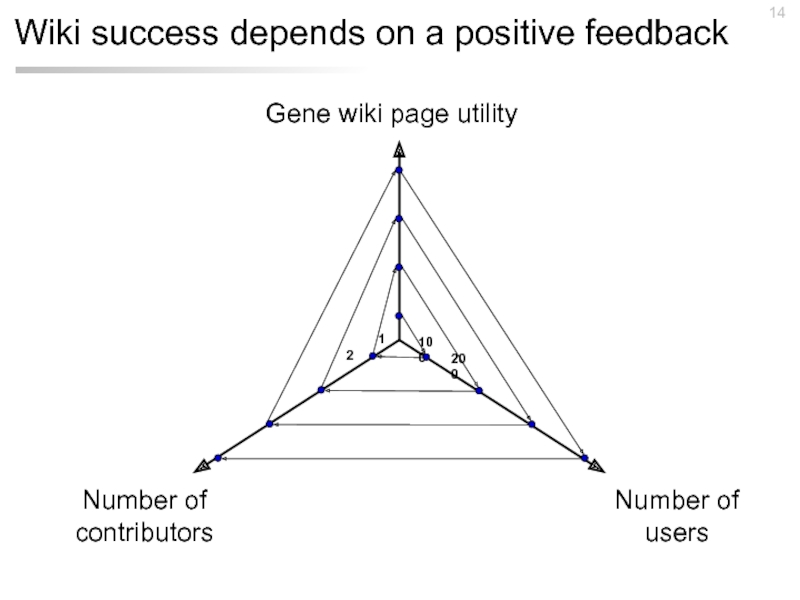
Слайд 14Wiki success depends on a positive feedback
Gene wiki page utility
Number of
users
Number
contributors
100
1
200
2
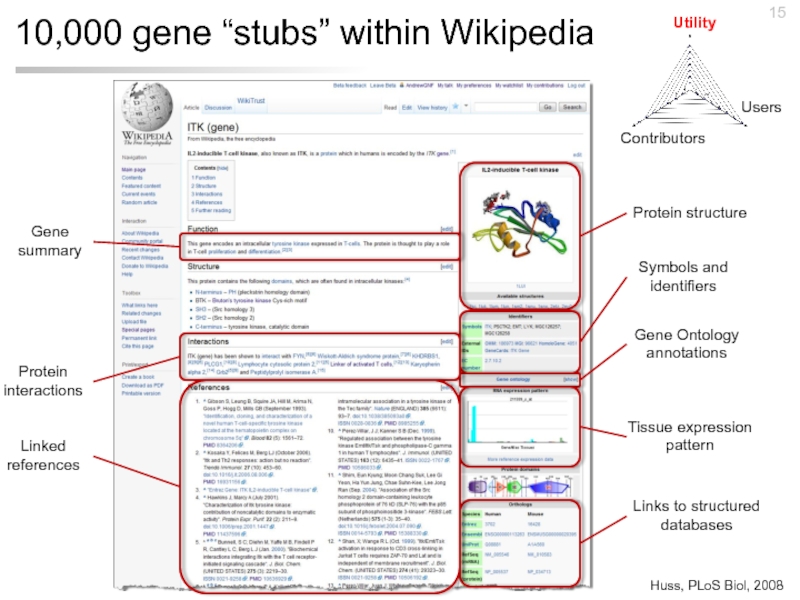
Слайд 1510,000 gene “stubs” within Wikipedia
Protein structure
Symbols and identifiers
Tissue expression pattern
Gene Ontology
Links to structured databases
Gene summary
Protein interactions
Linked references
Huss, PLoS Biol, 2008
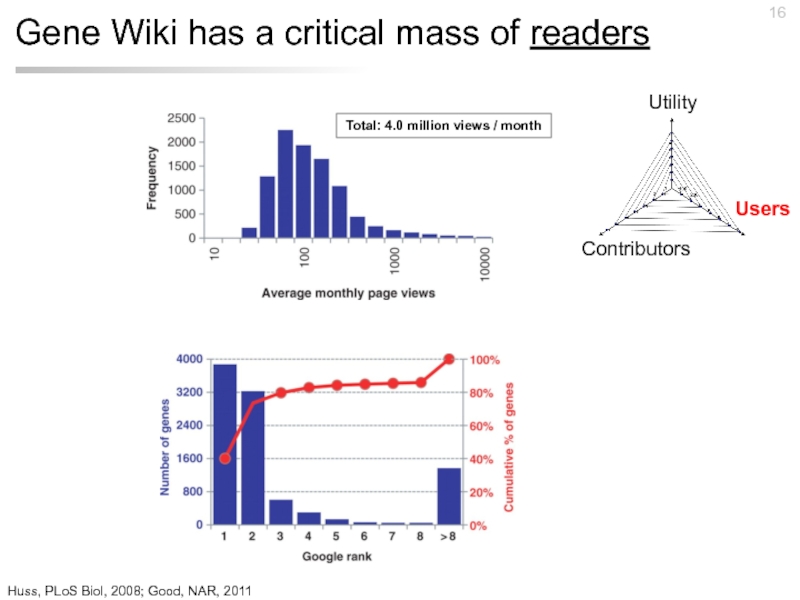
Слайд 16Gene Wiki has a critical mass of readers
Total: 4.0 million views
Huss, PLoS Biol, 2008; Good, NAR, 2011
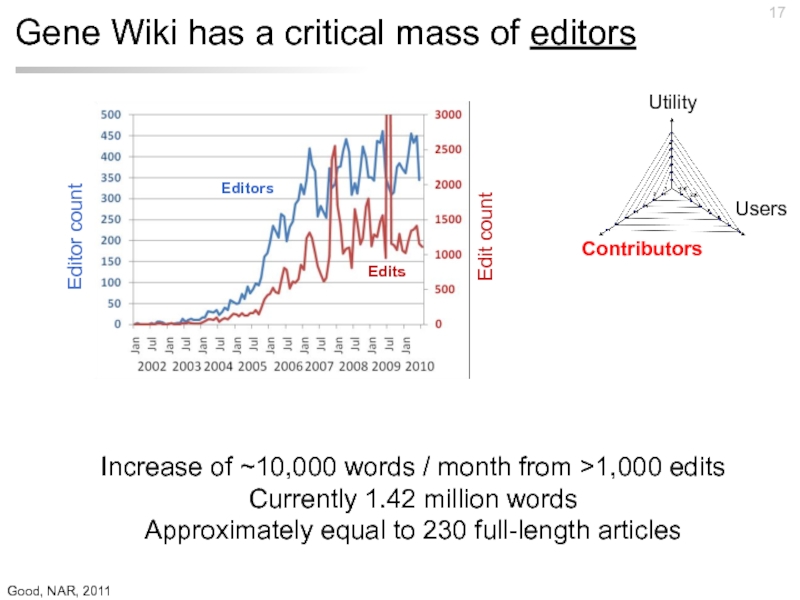
Слайд 17Gene Wiki has a critical mass of editors
Increase of ~10,000 words
Currently 1.42 million words
Approximately equal to 230 full-length articles
Good, NAR, 2011
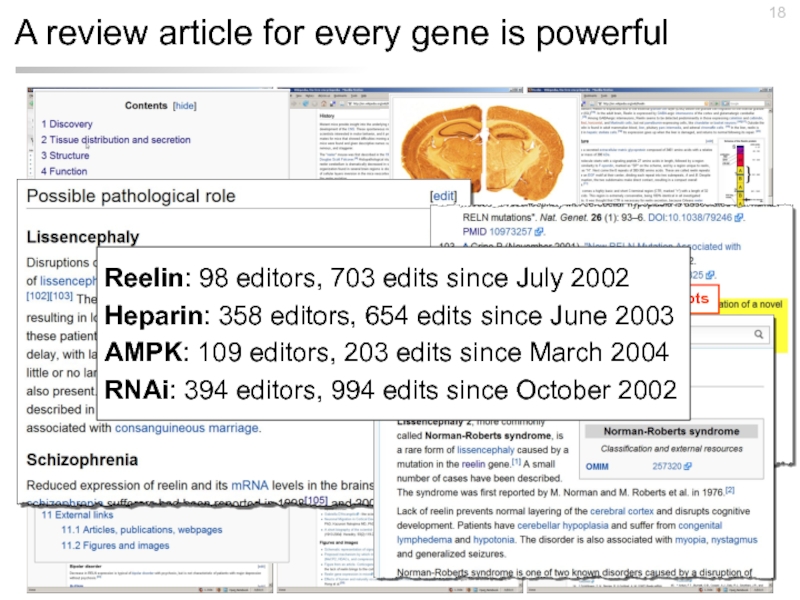
Слайд 18A review article for every gene is powerful
References to the literature
Hyperlinks
Reelin: 98 editors, 703 edits since July 2002
Heparin: 358 editors, 654 edits since June 2003
AMPK: 109 editors, 203 edits since March 2004
RNAi: 394 editors, 994 edits since October 2002
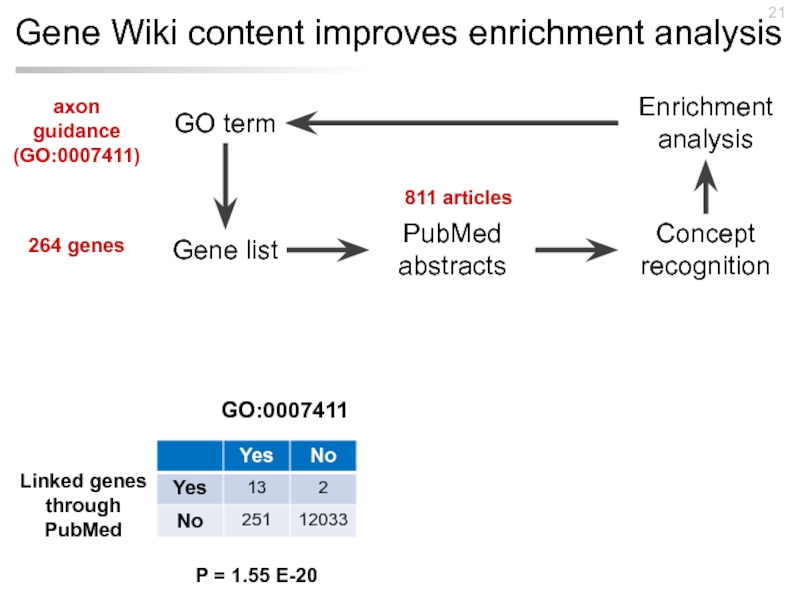
Слайд 21Gene Wiki content improves enrichment analysis
GO term
Gene list
Concept recognition
PubMed abstracts
Enrichment analysis
GO:0007411
axon
(GO:0007411)
264 genes
Linked genes through PubMed
P = 1.55 E-20
811 articles
Слайд 22Gene Wiki content improves enrichment analysis
GO term
Gene list
Concept recognition
PubMed abstracts
Enrichment analysis
GO:0006936
GO:0006936
muscle
87 genes
Linked genes through PubMed
Linked genes through PubMed + Gene Wiki
P = 1.0
P = 1.22 E-09
251 articles
87 articles
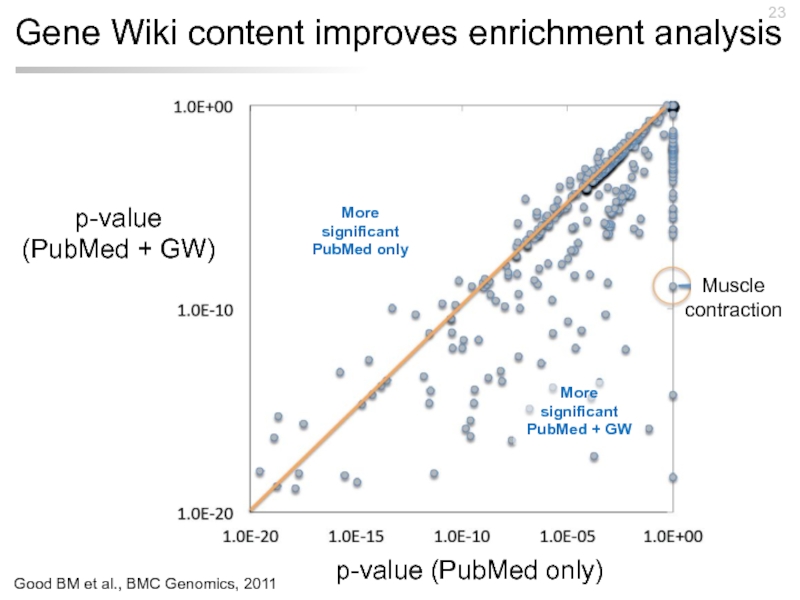
Слайд 23Gene Wiki content improves enrichment analysis
p-value (PubMed only)
p-value (PubMed + GW)
Muscle
More significant PubMed + GW
More significant PubMed only
Good BM et al., BMC Genomics, 2011
Слайд 27Cardiovascular Gene Wiki Portal
CAMK2D -- CaM kinase II subunit delta
CSRP3 --
GJA1 -- Gap junction alpha-1 protein / Connexin-43
MAPK14 -- Mitogen-activated protein kinase 14 / p38-α
MYL7 -- Myosin regulatory light chain 2, atrial isoform
MYL2 -- Myosin regulatory light chain 2, ventricular/cardiac isoform
PECAM1 -- Platelet endothelial cell adhesion molecule/CD31
RYR2 -- Ryanodine receptor 2
ATP2A2 -- Sarcoplasmic/endoplasmic reticulum calcium ATPase 2 / SERCA2
TNNI3 -- Troponin I, cardiac muscle
TNNT2 -- Troponin T, cardiac muscle
Peipei Ping
UCLA
Слайд 31Why is there so much redundancy?
Users
Requests
Resources
Time
Community
development
BioGPS emphasizes community extensibility
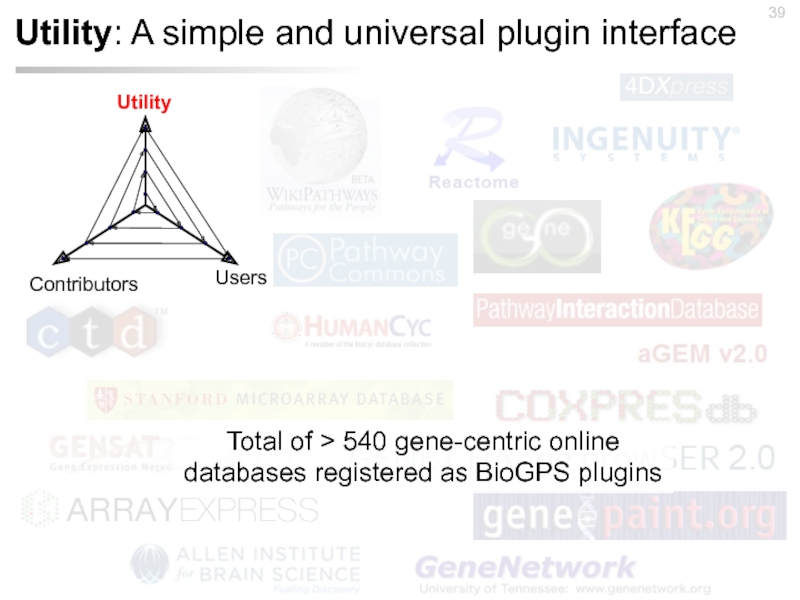
Слайд 39Utility: A simple and universal plugin interface
Total of > 540 gene-centric
Слайд 41Contributors: Explicit and implicit knowledge
540 plugins registered
(>300 publicly shared)
by over
spanning 280+ domains
Слайд 42Gene Annotation Query as a Service
http://mygene.info
High performance
3M hits/month
Highly scalable
13k species
16M genes
Weekly
JSON output
REST interface
Python/R/JS libraries
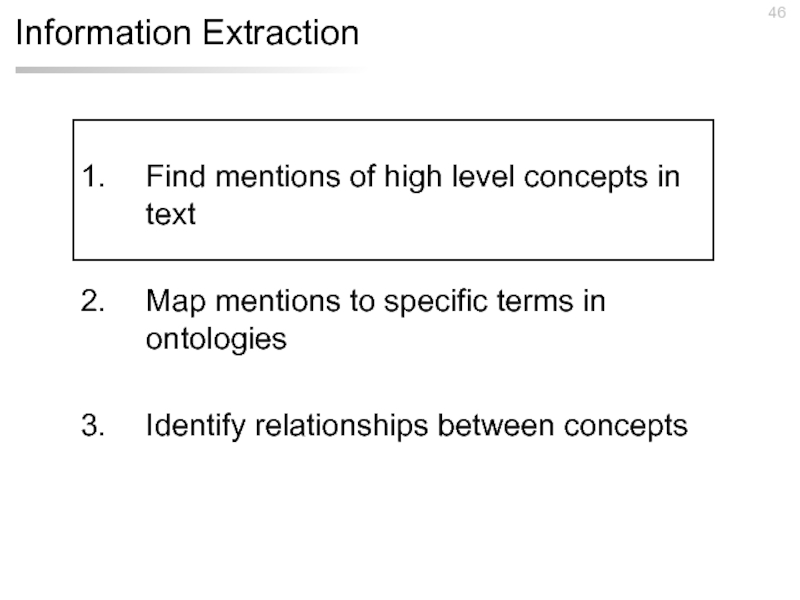
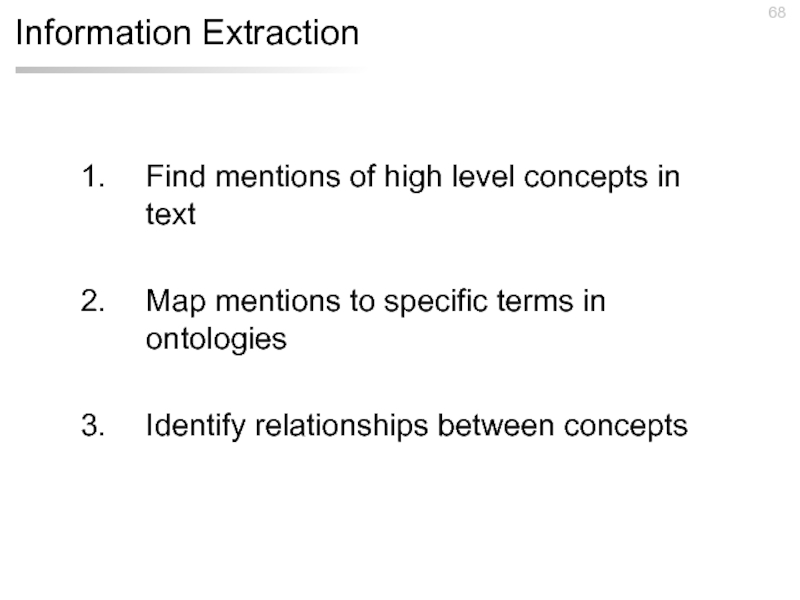
Слайд 46Information Extraction
Find mentions of high level concepts in text
Map mentions to
Identify relationships between concepts
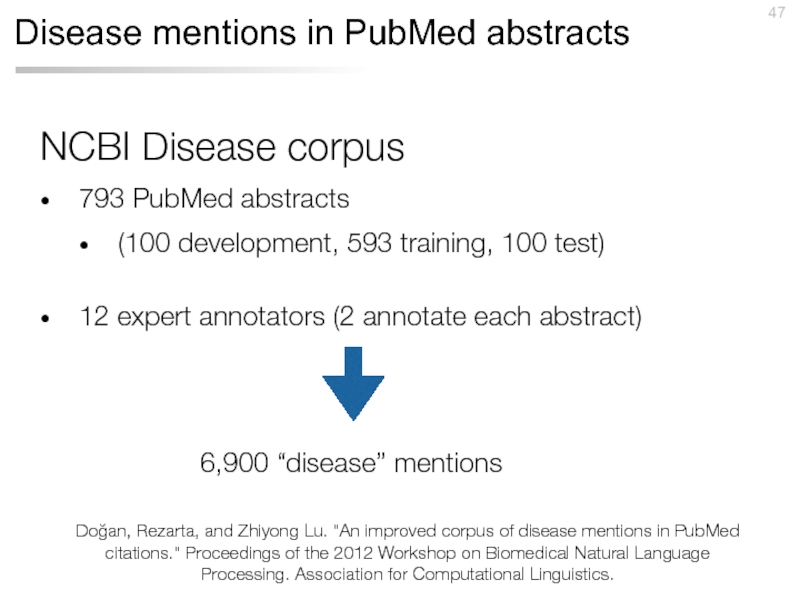
Слайд 47Disease mentions in PubMed abstracts
NCBI Disease corpus
793 PubMed abstracts
(100 development,
12 expert annotators (2 annotate each abstract)
6,900 “disease” mentions
Doğan, Rezarta, and Zhiyong Lu. "An improved corpus of disease mentions in PubMed citations." Proceedings of the 2012 Workshop on Biomedical Natural Language Processing. Association for Computational Linguistics.
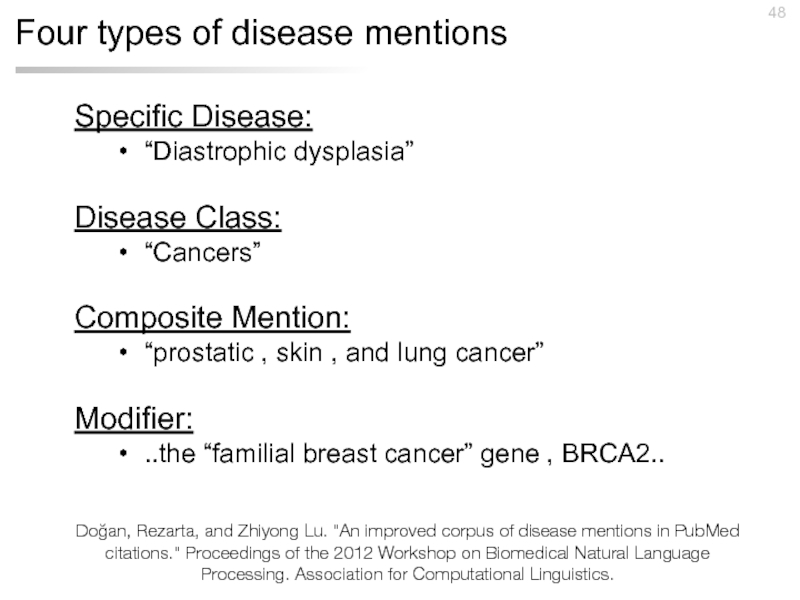
Слайд 48Four types of disease mentions
Specific Disease:
“Diastrophic dysplasia”
Disease Class:
“Cancers”
Composite Mention:
“prostatic , skin , and lung cancer”
Modifier:
..the “familial breast cancer” gene , BRCA2..
Doğan, Rezarta, and Zhiyong Lu. "An improved corpus of disease mentions in PubMed citations." Proceedings of the 2012 Workshop on Biomedical Natural Language Processing. Association for Computational Linguistics.
Слайд 49Question: Can a group of non-scientists collectively perform concept recognition in
Слайд 52Amazon Mechanical Turk (AMT)
For each task, specify:
a qualification test
how many workers
how much we will pay per task
Manages:
parallel execution of jobs
worker access to tasks via qualification tests
payments
task advertising
1. Create tasks
2. Execute
3. Aggregate
Слайд 53Instructions to workers
Highlight all diseases and disease abbreviations
“...are associated with
“The Wiskott-Aldrich syndrome ( WAS ) , an X-linked immunodeficiency…”
Highlight the longest span of text specific to a disease
“... contains the insulin-dependent diabetes mellitus locus …”
Highlight disease conjunctions as single, long spans.
“... a significant fraction of familial breast and ovarian cancer , but undergoes…”
Highlight symptoms - physical results of having a disease
“XFE progeroid syndrome can cause dwarfism, cachexia, and microcephaly. Patients often display learning disabilities, hearing loss, and visual impairment.
Слайд 54Qualification test
Test #1: “Myotonic dystrophy ( DM ) is associated with
Test #2: “Germline mutations in BRCA1 are responsible for most cases of inherited breast and ovarian cancer . However , the function of the BRCA1 protein has remained elusive . As a regulated secretory protein , BRCA1 appears to function by a mechanism not previously described for tumour suppressor gene products.”
Test #3: “We report about Dr . Kniest , who first described the condition in 1952 , and his patient , who , at the age of 50 years is severely handicapped with short stature , restricted joint mobility , and blindness but is mentally alert and leads an active life . This is in accordance with molecular findings in other patients with Kniest dysplasia and…”
26 yes / no questions
Слайд 57Experimental design
Task: Identify the disease mentions in the 593 abstracts from
$0.06 per Human Intelligence Task (HIT)
HIT = annotate one abstract from PubMed
5 workers annotate each abstract
Слайд 65Comparisons to human annotators
Average level of agreement between expert annotators (stage
F = 0.76
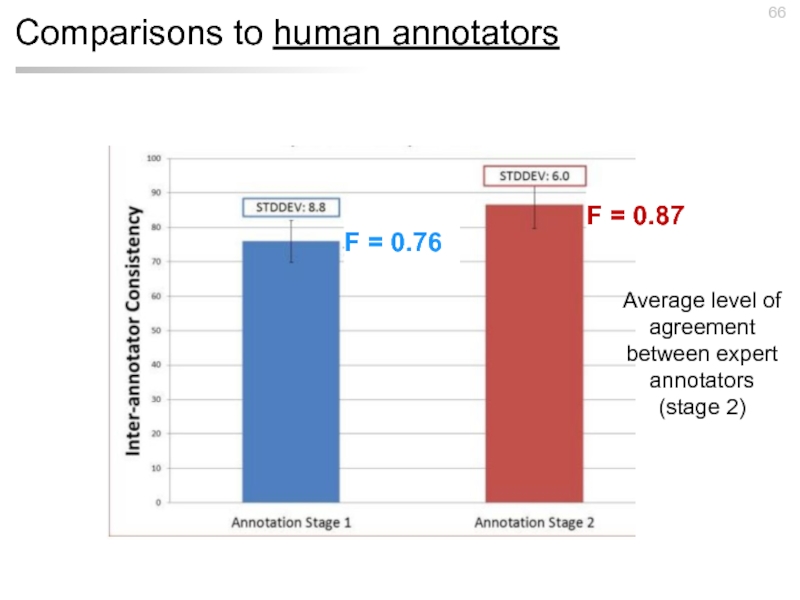
Слайд 66Comparisons to human annotators
F = 0.76
F = 0.87
Average level of agreement
(stage 2)
Слайд 67In aggregate, our worker ensemble is faster, cheaper and as accurate
Слайд 68Information Extraction
Find mentions of high level concepts in text
Map mentions to
Identify relationships between concepts
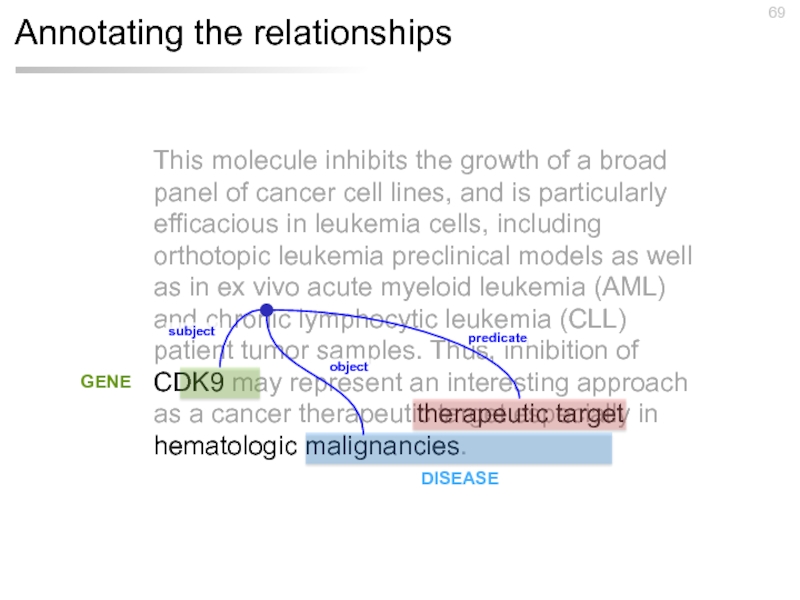
Слайд 69Annotating the relationships
This molecule inhibits the growth of a broad panel
therapeutic target
subject
predicate
object
GENE
DISEASE
Слайд 72Doug Howe, ZFIN
John Hogenesch, U Penn
Jon Huss, GNF
Luca de Alfaro, UCSC
Angel
Faramarz Valafar, SDSU
Pierre Lindenbaum,
Fondation Jean Dausset
Michael Martone, Rush
Konrad Koehler, Karo Bio
Warren Kibbe, Simon Lim, Northwestern
Lynn Schriml, U Maryland
Paul Pavlidis, U British Columbia
Peipei Ping, UCLA
Many Wikipedia editors
WP:MCB Project
Collaborators
Contact
http://sulab.org
asu@scripps.edu
@andrewsu
+Andrew Su
Citizen Science logo based on http://thenounproject.com/term/teamwork/39543/
Слайд 73Related AMT work
[1] Zhai et al 2013, used similar protocol to
[2] Burger et al, using microtask workers to identify relationships between genes and mutations.
[3] Aroyo & Welty, used workers to identify relations between concepts in medical text.
[1] Zhai H. et al (2013) ”Web 2.0-Based Crowdsourcing for High-Quality Gold Standard Development in Clinical Natural Language Processing” J Med Internet Res
[2] Burger, John, et al. (2014) "Hybrid curation of gene-mutation relations combining automated extraction and crowdsourcing.” Mitre technical report
[3] Aroyo, Lora, and Chris Welty. Harnessing disagreement in crowdsourcing a relation extraction gold standard. Tech. Rep. RC25371 (WAT1304-058), IBM Research, 2013.
























![Related AMT work[1] Zhai et al 2013, used similar protocol to tag medication names in](/img/tmb/4/309271/750a2a176f3a1bd1762890120d468f1a-800x.jpg)





