C. Titus Brown
ctb@msu.edu
May 21, 2014
- Главная
- Разное
- Дизайн
- Бизнес и предпринимательство
- Аналитика
- Образование
- Развлечения
- Красота и здоровье
- Финансы
- Государство
- Путешествия
- Спорт
- Недвижимость
- Армия
- Графика
- Культурология
- Еда и кулинария
- Лингвистика
- Английский язык
- Астрономия
- Алгебра
- Биология
- География
- Детские презентации
- Информатика
- История
- Литература
- Маркетинг
- Математика
- Медицина
- Менеджмент
- Музыка
- МХК
- Немецкий язык
- ОБЖ
- Обществознание
- Окружающий мир
- Педагогика
- Русский язык
- Технология
- Физика
- Философия
- Химия
- Шаблоны, картинки для презентаций
- Экология
- Экономика
- Юриспруденция
Six ways to Sunday: approaches to computational reproducibility in non-model system sequence analysis. презентация
Содержание
- 1. Six ways to Sunday: approaches to computational reproducibility in non-model system sequence analysis.
- 2. Hello! Assistant Professor; Microbiology; Computer Science; etc.
- 3. The challenges of non-model sequencing Missing or
- 4. Shotgun sequencing & assembly http://eofdreams.com/library.html; http://www.theshreddingservices.com/2011/11/paper-shredding-services-small-business/; http://schoolworkhelper.net/charles-dickens%E2%80%99-tale-of-two-cities-summary-analysis/
- 5. Shotgun sequencing analysis goals: Assembly (what is
- 6. Assembly It was the best of times,
- 7. Shared low-level fragments may not reach
- 8. Introducing k-mers CCGATTGCACTGGACCGA (
- 9. K-mers give you an implicit alignment
- 10. K-mers give you an implicit alignment
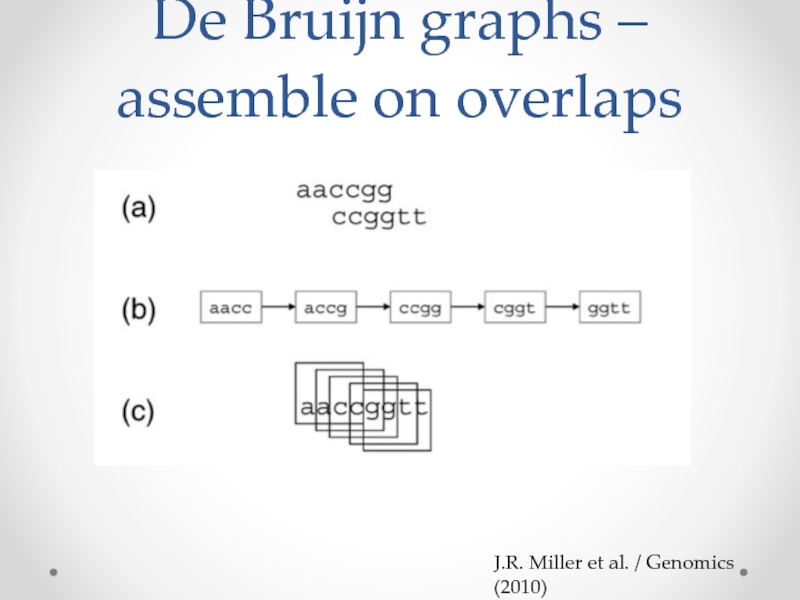
- 11. De Bruijn graphs – assemble on overlaps J.R. Miller et al. / Genomics (2010)
- 12. The problem with k-mers
- 13. Conway T C , Bromage A J
- 14. Practical memory measurements (soil) Velvet measurements (Adina Howe)
- 15. Data set size and cost $1000 gets
- 16. Efficient data structures & algorithms
- 17. Shotgun sequencing is massively redundant; can we
- 18. Sparse collections of k-mers can be stored
- 19. Data structures & algorithms papers “These
- 20. Data analysis papers “Tackling soil diversity
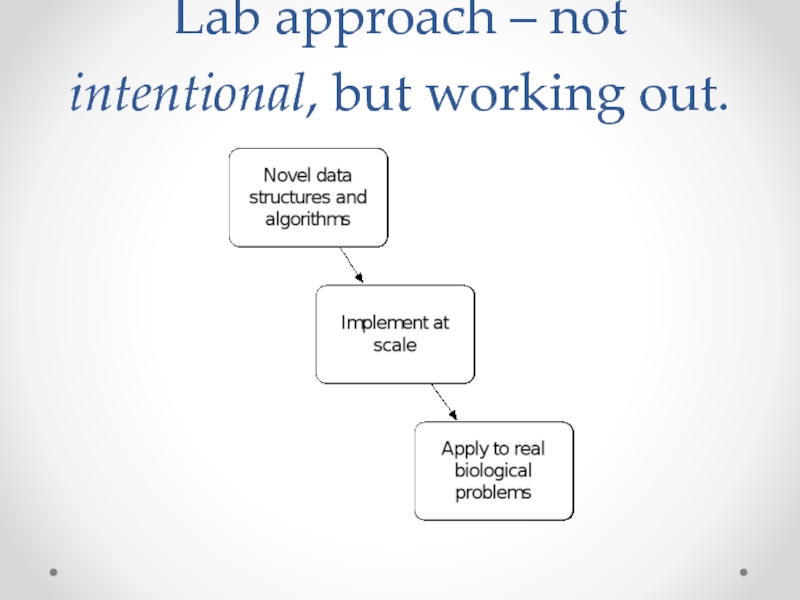
- 21. Lab approach – not intentional, but working out.
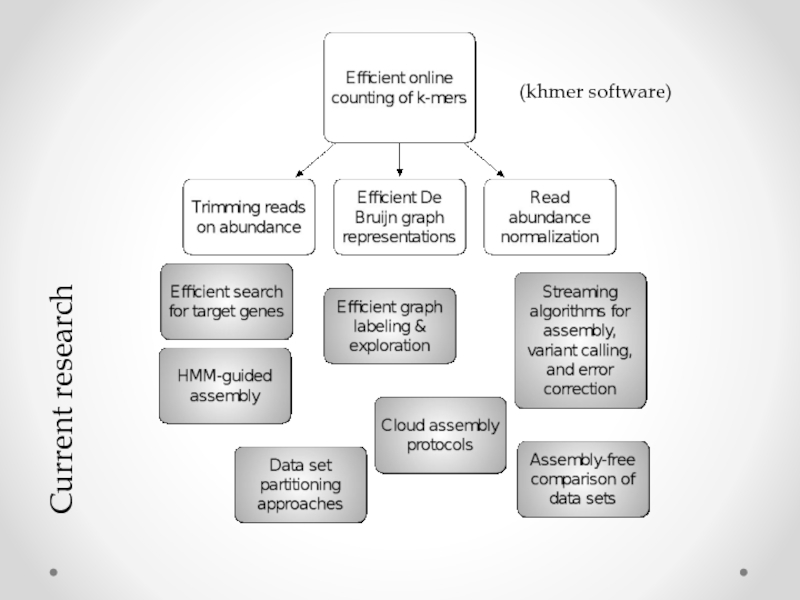
- 22. This leads to good things. (khmer software)
- 23. Current research (khmer software)
- 24. Testing & version control – the not
- 25. On the “novel research” side:
- 26. Running entirely w/in cloud Complete data; AWS
- 27. On the “novel research” side:
- 28. Reproducibility! Scientific progress relies on reproducibility
- 29. Disclaimer Not a researcher of reproducibility!
- 30. My usual intro: We practice open science!
- 31. My usual intro: We practice open science!
- 32. My lab & the diginorm paper. All
- 33. IPython Notebook: data + code =>
- 34. My lab & the diginorm paper. All
- 35. To reproduce our paper: git
- 36. Now standard in lab -- All our
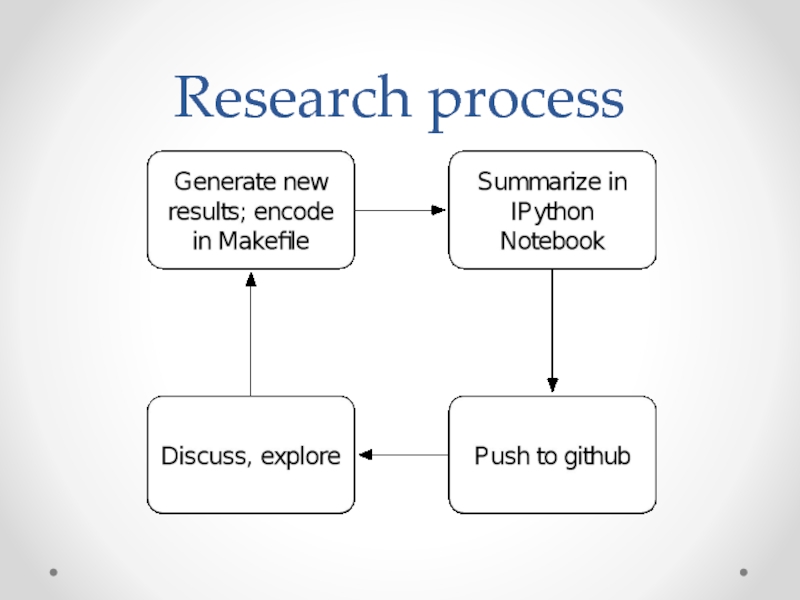
- 37. Research process
- 38. Literate graphing & interactive exploration
- 39. The process We start with pipeline
- 40. Growing & refining the process Now
- 41. 1. Use standard OS; provide install instructions
- 42. 2. Automate Literate graphing now easy
- 43. Myths of reproducible research (Opinions from personal experience.)
- 44. Myth 1: Partial reproducibility is hard.
- 45. Myth 2: Incomplete reproducibility is useless
- 46. Myth 3: We need new platforms
- 47. Myth 4. Virtual Machine reproducibility is an
- 48. Myth 5: We can use GUIs for
- 49. Our current efforts? Semantic versioning of
- 50. khmer-protocols
- 51. khmer-protocols: Provide standard “cheap” assembly protocols for
- 52. Literate testing Our shell-command tutorials for bioinformatics
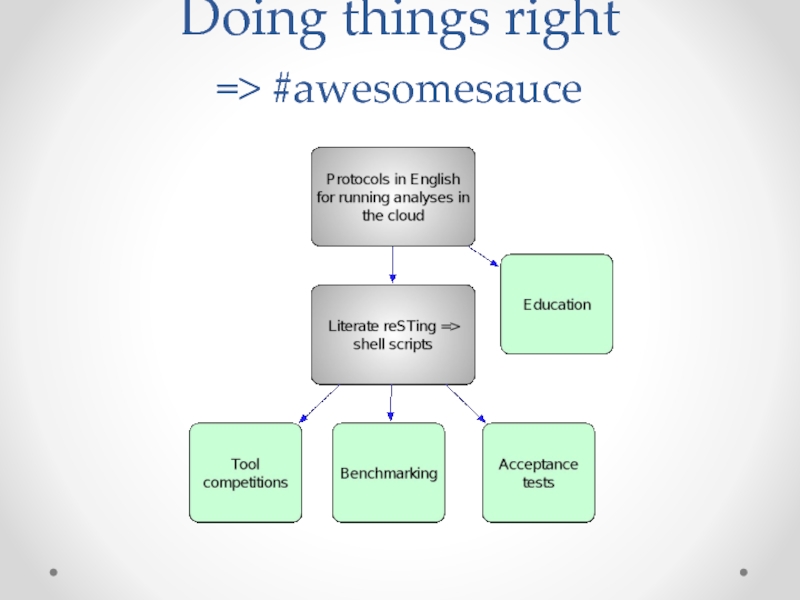
- 53. Doing things right => #awesomesauce
- 54. Concluding thoughts We are not doing anything
- 55. What bits should people adopt? Version
- 56. More concluding thoughts Nobody would care
- 57. Biology & sequence analysis is in a
- 58. Thanks! Talk is on slideshare: slideshare.net/c.titus.brown E-mail or tweet me: ctb@msu.edu @ctitusbrown
Слайд 1Six ways to Sunday: approaches to computational reproducibility in non-model system
Слайд 2Hello!
Assistant Professor; Microbiology; Computer Science; etc.
More information at:
ged.msu.edu/
github.com/ged-lab/
ivory.idyll.org/blog/
@ctitusbrown
Слайд 3The challenges of non-model sequencing
Missing or low quality genome reference.
Evolutionarily distant.
Most
Assume low polymorphism (internal variation)
Assume reference genome
Assume somewhat reliable functional annotation
More significant compute infrastructure
…and cannot easily or directly be used on critters of interest.
Слайд 4Shotgun sequencing & assembly
http://eofdreams.com/library.html;
http://www.theshreddingservices.com/2011/11/paper-shredding-services-small-business/;
http://schoolworkhelper.net/charles-dickens%E2%80%99-tale-of-two-cities-summary-analysis/
Слайд 5Shotgun sequencing analysis goals:
Assembly (what is the text?)
Produces new genomes &
Gene discovery for enzymes, drug targets, etc.
Counting (how many copies of each book?)
Measure gene expression levels, protein-DNA interactions
Variant calling (how does each edition vary?)
Discover genetic variation: genotyping, linkage studies…
Allele-specific expression analysis.
Слайд 6Assembly
It was the best of times, it was the wor
, it
isdom, it was the age of foolishness
mes, it was the age of wisdom, it was th
It was the best of times, it was the worst of times, it was the age of wisdom, it was the age of foolishness
…but for lots and lots of fragments!
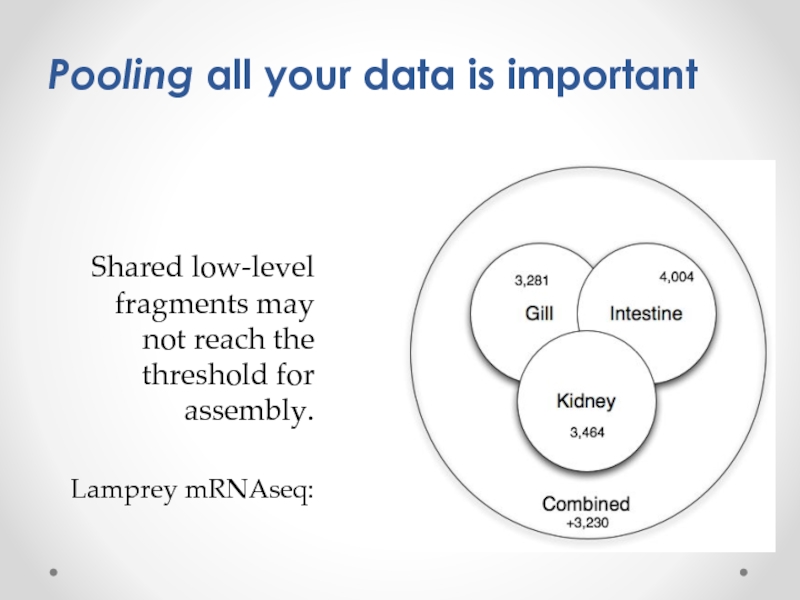
Слайд 7
Shared low-level fragments may not reach the threshold for assembly.
Lamprey mRNAseq:
Pooling
Слайд 9K-mers give you an implicit alignment
CCGATTGCACTGGACCGATGCACGGTACCGTATAGCC
CATGGACCGATTGCACTGGACCGATGCACGGTACCG
Слайд 10K-mers give you an implicit alignment
CCGATTGCACTGGACCGATGCACGGTACCGTATAGCC
CATGGACCGATTGCACTGGACCGATGCACGGTACCG
CATGGACCGATTGCACTGGACCGATGCACGGACCG
(with no accounting for mismatches or indels)
Слайд 12The problem with k-mers
CCGATTGCACTGGACCGATGCACGGTACCGTATAGCC
CATGGACCGATTGCACTCGACCGATGCACGGTACCG
Each sequencing
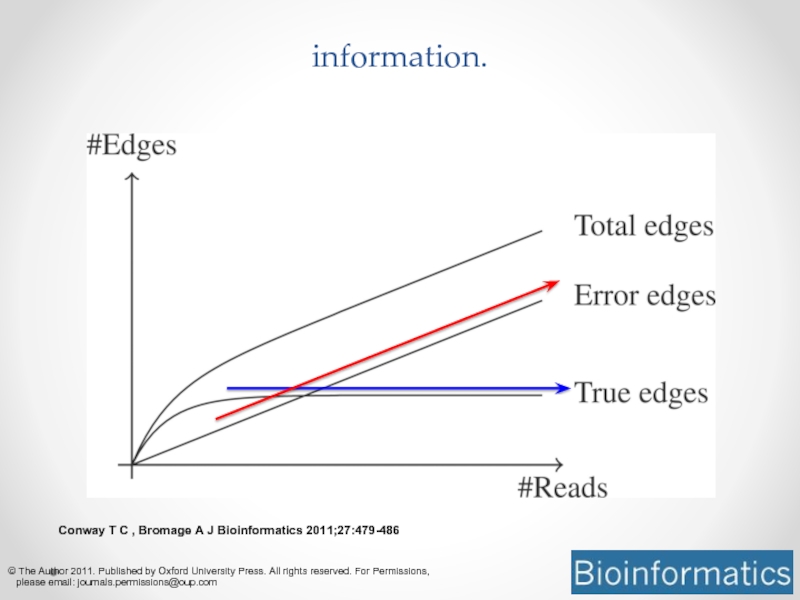
Слайд 13Conway T C , Bromage A J Bioinformatics 2011;27:479-486
© The Author
Assembly graphs scale with data size, not information.
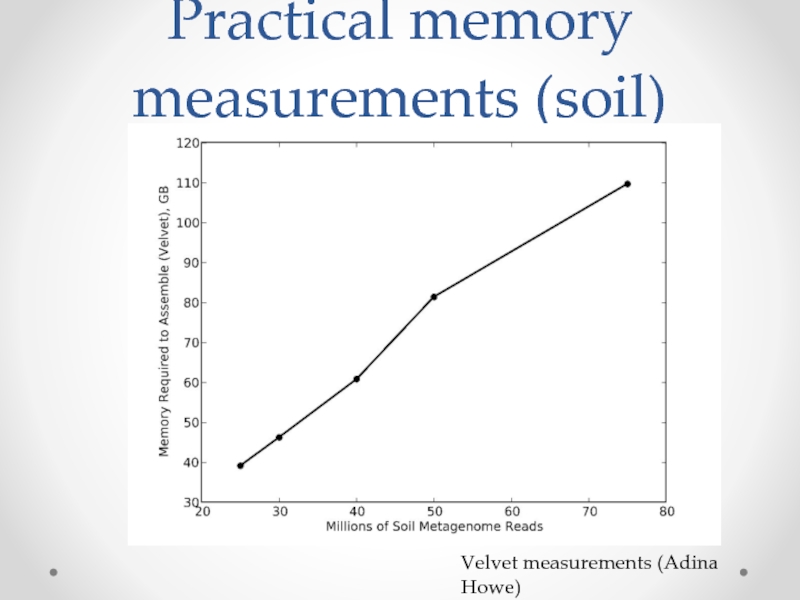
Слайд 15Data set size and cost
$1000 gets you ~100m “reads”, or about
> 1000 labs doing this regularly.
Each data set analysis is ~custom.
Analyses are data intensive and memory intensive.
Слайд 17Shotgun sequencing is massively redundant; can we eliminate redundancy while retaining
Analog: JPEG lossy compression
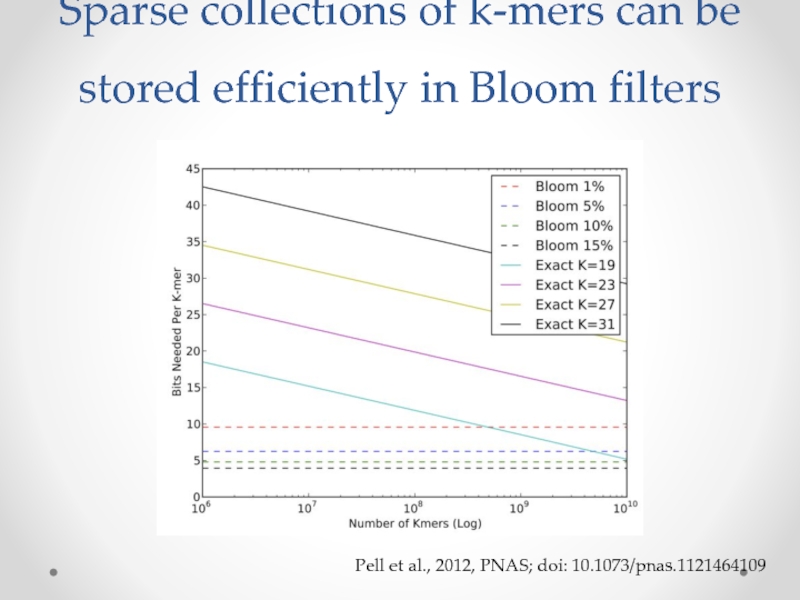
Слайд 18Sparse collections of k-mers can be stored efficiently in Bloom filters
Pell
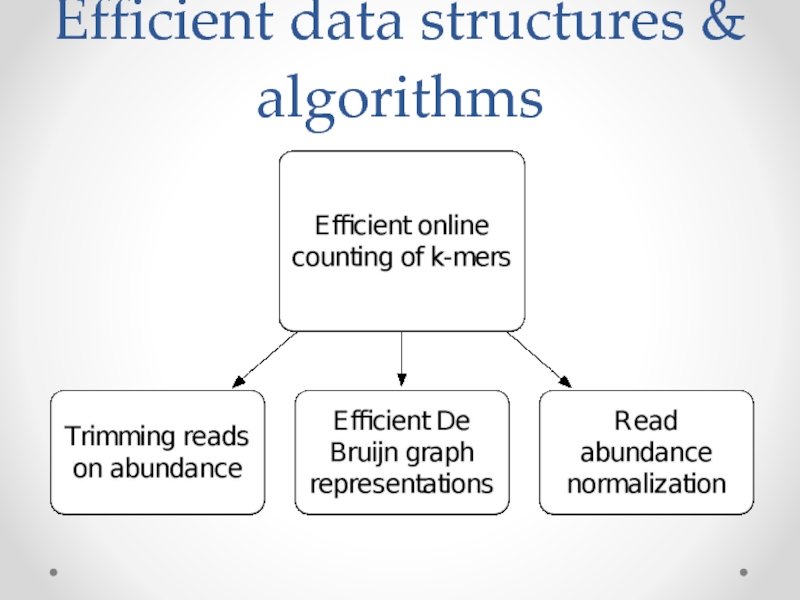
Слайд 19Data structures & algorithms papers
“These are not the k-mers you are
“Scaling metagenome sequence assembly with probabilistic de Bruijn graphs”, Pell et al., PNAS 2012.
“A Reference-Free Algorithm for Computational Normalization of Shotgun Sequencing Data”, Brown et al., arXiv 1203.4802, under revision.
Слайд 20Data analysis papers
“Tackling soil diversity with the assembly of large, complex
Assembling novel ascidian genomes & transcriptomes, Lowe et al., in prep.
A de novo lamprey transcriptome from large scale multi-tissue mRNAseq, Scott et al., in prep.
Слайд 24Testing & version control – the not so secret sauce
High test
Stupidity driven testing – we write tests for bugs after we find them and before we fix them.
Pull requests & continuous integration – does your proposed merge break tests?
Pull requests & code review – does new code meet our minimal coding etc requirements?
Note: spellchecking!!!
Слайд 25On the “novel research” side:
Novel data structures and algorithms;
Permit low(er) memory
Liberate analyses from specialized hardware.
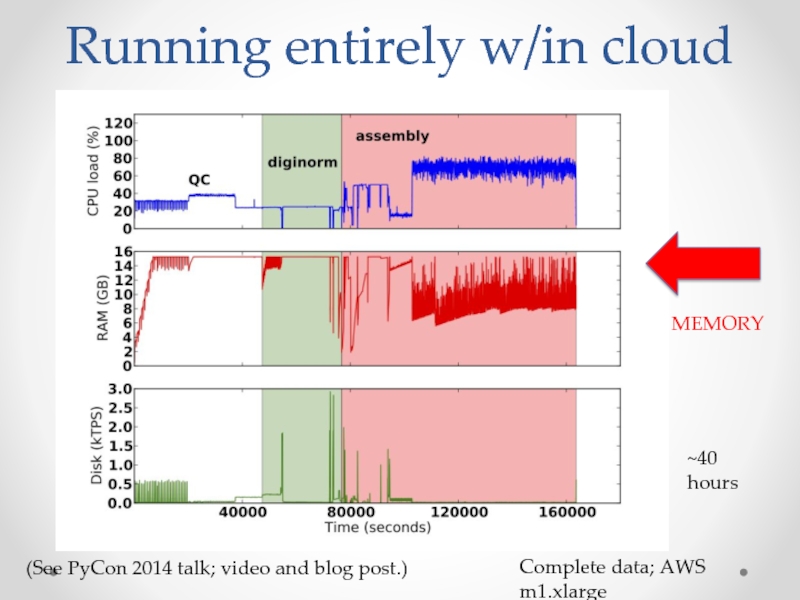
Слайд 26Running entirely w/in cloud
Complete data; AWS m1.xlarge
~40 hours
(See PyCon 2014 talk;
MEMORY
Слайд 27On the “novel research” side:
Novel data structures and algorithms;
Permit low(er) memory
Liberate analyses from specialized hardware.
This last bit? => reproducibility.
Слайд 28Reproducibility!
Scientific progress relies on reproducibility of analysis. (Aristotle, Nature, 322 BCE.)
“There
Слайд 29Disclaimer
Not a researcher of reproducibility!
Merely a practitioner.
Please take my points below
(But I’m right.)
Слайд 30My usual intro:
We practice open science!
Everything discussed here:
Code: github.com/ged-lab/ ; BSD
Blog: http://ivory.idyll.org/blog (‘titus brown blog’)
Twitter: @ctitusbrown
Grants on Lab Web site: http://ged.msu.edu/research.html
Preprints available.
Everything is > 80% reproducible.
Слайд 31My usual intro:
We practice open science!
Everything discussed here:
Code: github.com/ged-lab/ ; BSD
Blog: http://ivory.idyll.org/blog (‘titus brown blog’)
Twitter: @ctitusbrown
Grants on Lab Web site: http://ged.msu.edu/research.html
Preprints available.
Everything is > 80% reproducible.
Слайд 32My lab & the diginorm paper.
All our code was already on
Much of our data analysis was already in the cloud;
Our figures were already made in IPython Notebook
Our paper was already in LaTeX
Слайд 34My lab & the diginorm paper.
All our code was already on
Much of our data analysis was already in the cloud;
Our figures were already made in IPython Notebook
Our paper was already in LaTeX
…why not push a bit more and make it easily reproducible?
This involved writing a tutorial. And that’s it.
Слайд 35To reproduce our paper:
git clone && python setup.py install
git clone
cd pipeline
wget && tar xzf
make && cd ../notebook && make
cd ../ && make
Слайд 36Now standard in lab --
All our papers now have:
Source hosted on
Data hosted there or on AWS;
Long running data analysis => ‘make’
Graphing and data digestion => IPython Notebook (also in github)
Qingpeng Zhang
Слайд 39The process
We start with pipeline reproducibility
Baked into lab culture; default “use
Community of practice!
Use standard open source approaches, so OSS developers learn it easily.
Enables easy collaboration w/in lab
Valuable learning tool!
Слайд 40Growing & refining the process
Now moving to Ubuntu Long-Term Support +
Everything is as automated as is convenient.
Students expected to communicate with me in IPython Notebooks.
Trying to avoid building (or even using) new tools.
Avoid maintenance burden as much as possible.
Слайд 411. Use standard OS; provide install instructions
Providing install, execute for Ubuntu
Avoid pre-configured virtual machines!
Locks you into specific cloud homes.
Challenges remixability and extensibility.
Слайд 422. Automate
Literate graphing now easy with knitr and IPython Notebook.
Build automation
Explicit is better than implicit. Make it easy to understand what you’re doing and how to extend it.
Слайд 44Myth 1: Partial reproducibility is hard.
“Here’s my script.” => Methods
More
Many scientists cannot replicate any part of their analysis without a lot of manual work.
Automating this is a win for reasons that have nothing to do with reproducibility… efficiency!
See: Software Carpentry.
Слайд 45Myth 2: Incomplete reproducibility is useless
Paraphrase: “We can’t possibly reproduce the
(Analogous arg re software testing & code coverage.)
…I really have a hard time arguing the paraphrase honestly…
Being able to reanalyze your raw data? Interesting.
Knowing how you made your figures? Really useful.
Слайд 46Myth 3: We need new platforms
Techies always want to build something
We probably do need new platforms, but stop thinking that building them does a service.
Platforms need to be use driven. Seriously.
If you write good software for scientific inquiry and make it easy to use reproducibly, that will drive virtuousity.
Слайд 47Myth 4. Virtual Machine reproducibility is an end solution.
Good start! Better
But:
Limits understanding & reuse.
Limits remixing: often cannot install other software!
“Chinese Room” argument: could be just a lookup table.
Слайд 48Myth 5: We can use GUIs for reproducible research
(OK, this is
Almost all data analysis takes place within a larger pipeline; the GUI must consume entire pipeline in order to be reproducible.
IFF GUI wraps command line, that’s a decent compromise (e.g. Galaxy) but handicaps researchers using novel approaches.
By the time it’s in a GUI, it’s no longer research.
Слайд 49Our current efforts?
Semantic versioning of our own code: stable command-line interface.
Writing
Automate ‘em for testing purposes.
Encourage their use, inclusion, and adaptation by others.
Слайд 51khmer-protocols:
Provide standard “cheap” assembly protocols for the cloud.
Entirely copy/paste; ~2-6 days
Open, versioned, forkable, citable….
Слайд 52Literate testing
Our shell-command tutorials for bioinformatics can now be executed in
See: github.com/ged-lab/literate-resting/.
Tremendously improves peace of mind and confidence moving forward!
Leigh Sheneman
Слайд 54Concluding thoughts
We are not doing anything particularly neat on the computational
Much of our effort is now driven by sheer utility:
Automation reduces our maintenance burden.
Extensibility makes revisions much easier!
Explicit instructions are good for training.
Some effort needed at the beginning, but once practices are established, “virtuous cycle” takes over.
Слайд 55What bits should people adopt?
Version control!
Literate graphing!
Automated “build” from data =>
Make available data as early in your pipeline as possible.
Слайд 56More concluding thoughts
Nobody would care that we were doing things reproducibly
Make sure students realize that faffing about on infrastructure isn’t science.
Research is about doing science. Reproducibility (like other good practices) is much easier to proselytize if you can link it to progress in science.
Слайд 57Biology & sequence analysis is in a perfect place for reproducibility
We
Big Data: laptops are too small;
Excel doesn’t scale any more;
Few tools in use; most of them are $$ or UNIX;
Little in the way of entrenched research practice;